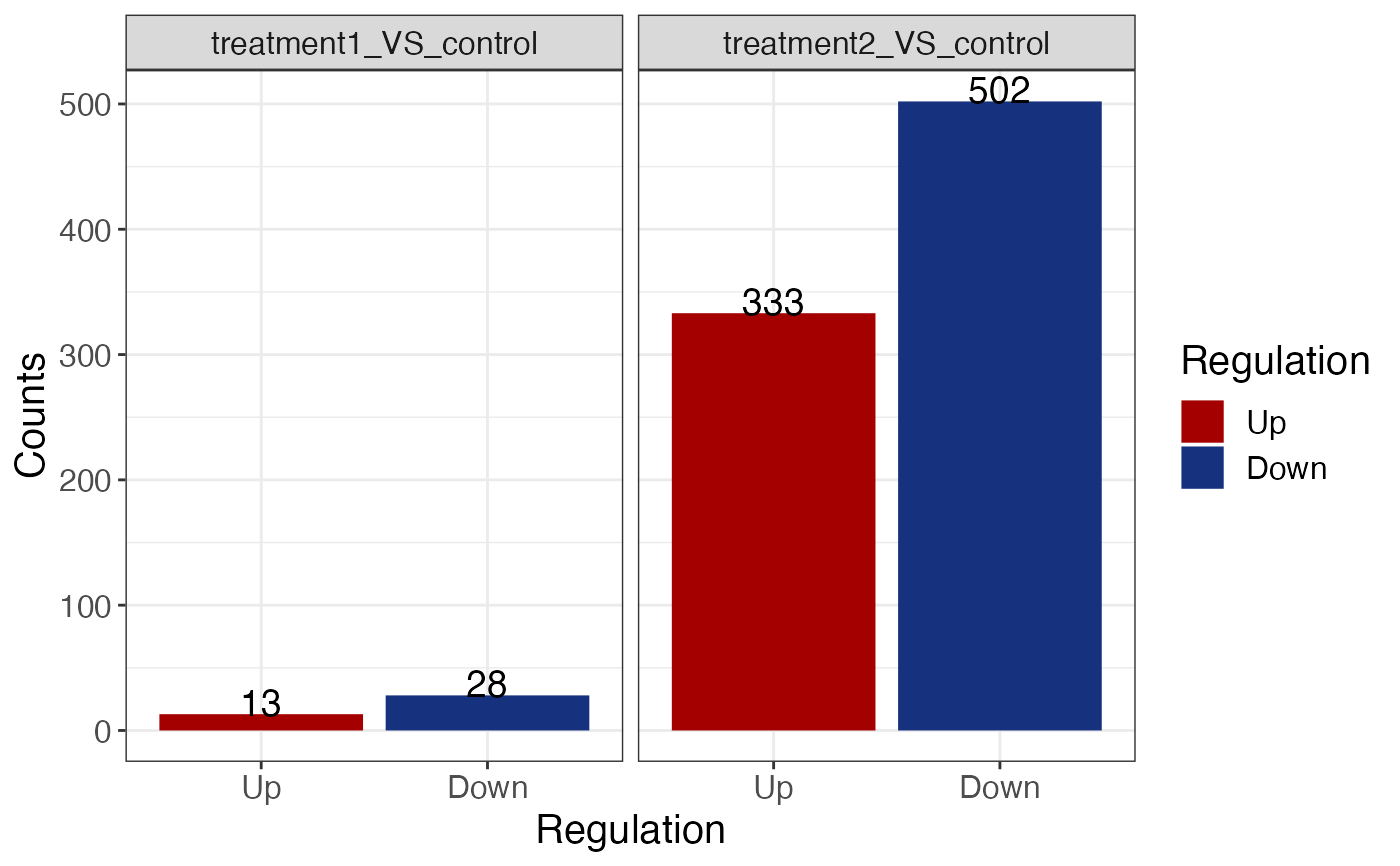
Generate a barplot of DEG counts.
Usage
get_diff_gene_count_barplot(
x,
col_up = "#a40000",
col_down = "#16317d",
font_size = 12,
show_counts = TRUE
)Arguments
- x
an object of class parcutils
- col_up
a character string, default #a40000, denoting valid a color name for "Up" regulated genes.
- col_down
a character string, default #16317d, denoting valid a color name "Down" regulated genes.
- font_size
a numeric, default 12, denoting font size in the plot.
- show_counts
a logical, default
TRUEdenoting whether to show counts on each bar.
Examples
res = .get_parcutils_object_example()
#> ℹ Running DESeq2 ...
#> converting counts to integer mode
#> Warning: some variables in design formula are characters, converting to factors
#> estimating size factors
#> estimating dispersions
#> gene-wise dispersion estimates
#> mean-dispersion relationship
#> final dispersion estimates
#> fitting model and testing
#> ✔ Done.
#> ℹ Summarizing DEG ...
#> ✔ Done.
get_diff_gene_count_barplot(res, font_size = 15)