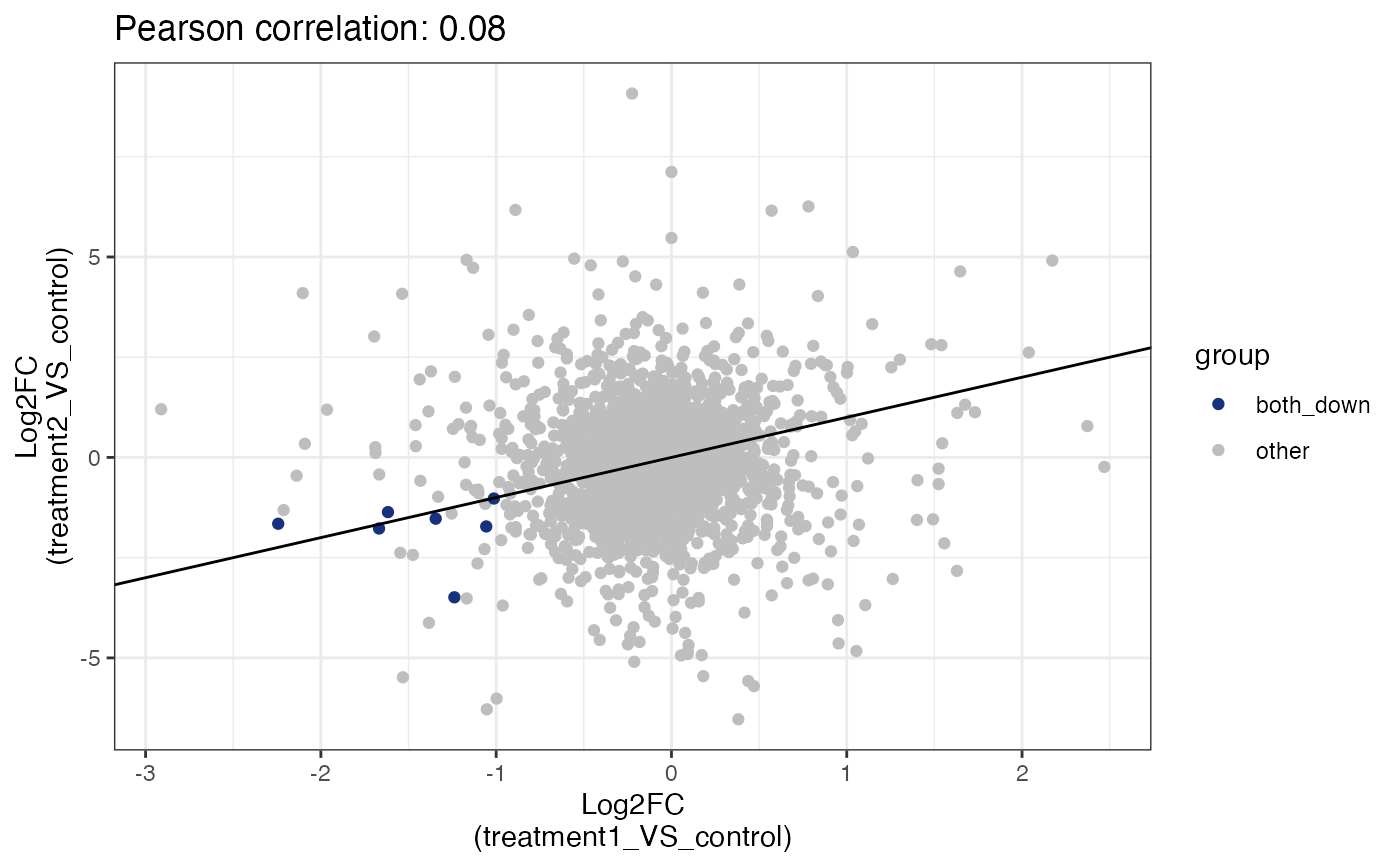
Compare log2 fold change between two sample comparisons.
Source:R/rnaseq_related.R
get_fold_change_scatter_plot.RdThis function generates a scatter plot of log2 fold change values for two different comparisons.
Usage
get_fold_change_scatter_plot(
x,
sample_comparisons,
labels = NULL,
point_size = 2,
label_size = 2,
color_label = "both",
col_up = "#a40000",
col_down = "#16317d",
show_diagonal_line = TRUE,
show_correlation = TRUE,
repair_genes = TRUE
)Arguments
- x
an object of class parcutils.
- sample_comparisons
a character vector of length 2 denoting sample comparisons to plot.
- labels
a character vector of genes to label. Default NULL, no labels.
- point_size
a numeric, default 2, denoting size of the points.
- label_size
a numeric, default 2, denoting size of the labels.
- color_label
a character string one of the "both", #both_down, or "both_up". Default
both.- col_up
a character string, default
#a40000, a valid color code for common up regulated genes.- col_down
a character string, default
#16317d, a valid color code for common down regulated genes.- show_diagonal_line
a logical, default TRUE, denoting whether to show a diagonal line.
- show_correlation
a logical, default TRUE, denoting whether to show Pearson correlation value.
- repair_genes
a logical, default
TRUE, denotes whether to repair gene names or not, IfTRUEstring prior to:will be removed from the gene names.
Examples
count_file <- system.file("extdata","toy_counts.txt" , package = "parcutils")
count_data <- readr::read_delim(count_file, delim = "\t")
#> Rows: 5000 Columns: 10
#> ── Column specification ────────────────────────────────────────────────────────
#> Delimiter: "\t"
#> chr (1): gene_id
#> dbl (9): control_rep1, control_rep2, control_rep3, treat1_rep1, treat1_rep2,...
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
sample_info <- count_data %>% colnames() %>% .[-1] %>%
tibble::tibble(samples = . , groups = rep(c("control" ,"treatment1" , "treatment2"), each = 3) )
res <- run_deseq_analysis(counts = count_data ,
sample_info = sample_info,
column_geneid = "gene_id" ,
group_numerator = c("treatment1", "treatment2") ,
group_denominator = c("control"))
#> ℹ Running DESeq2 ...
#> converting counts to integer mode
#> Warning: some variables in design formula are characters, converting to factors
#> estimating size factors
#> estimating dispersions
#> gene-wise dispersion estimates
#> mean-dispersion relationship
#> final dispersion estimates
#> fitting model and testing
#> ✔ Done.
#> ℹ Summarizing DEG ...
#> ✔ Done.
# show common up and common down
get_fold_change_scatter_plot(x = res,
sample_comparisons = c("treatment1_VS_control",
"treatment2_VS_control"),label_size = 3)
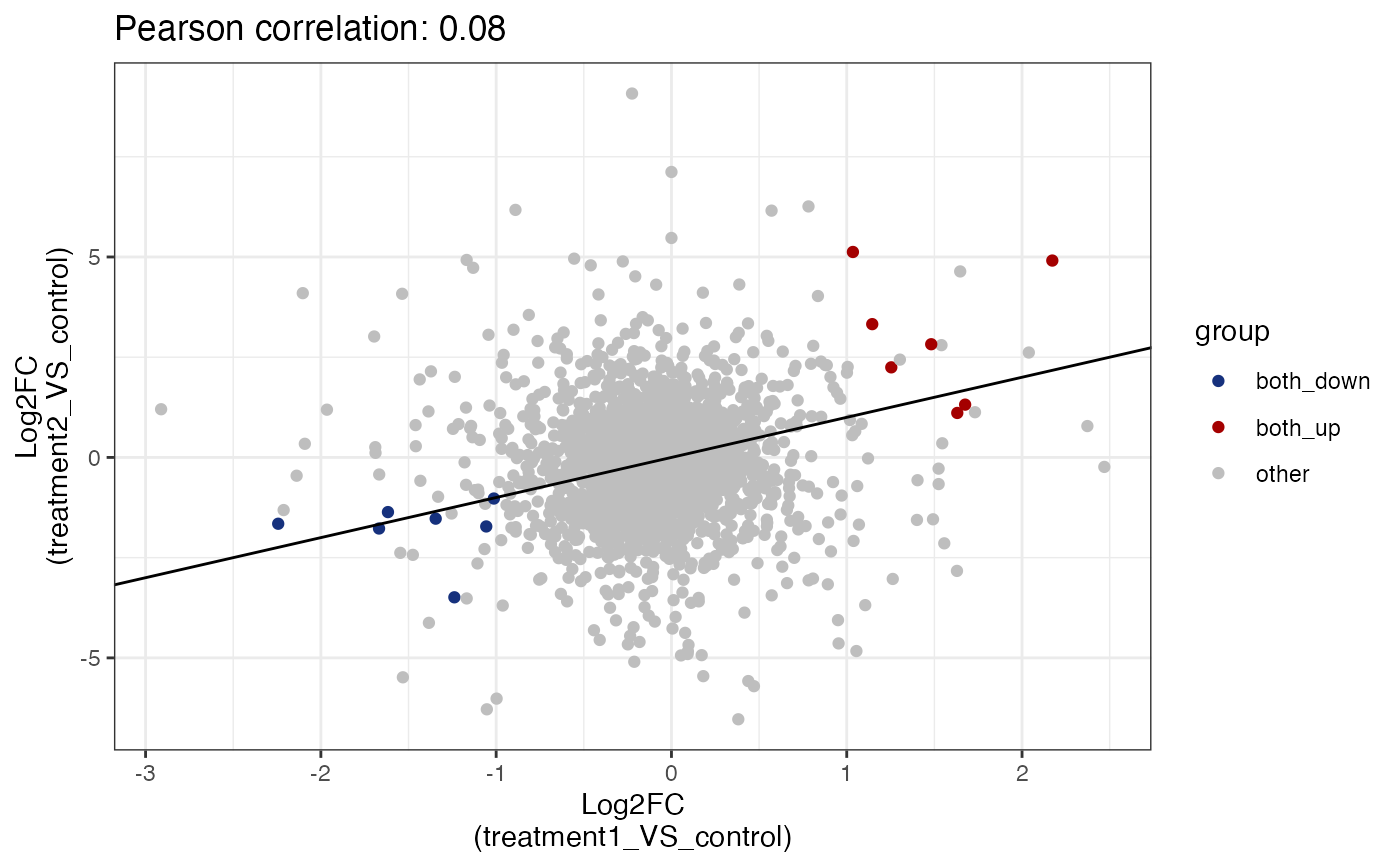 # show common up
get_fold_change_scatter_plot(x = res,
sample_comparisons = c("treatment1_VS_control",
"treatment2_VS_control"),
color_label = "both_up",label_size = 4)
# show common up
get_fold_change_scatter_plot(x = res,
sample_comparisons = c("treatment1_VS_control",
"treatment2_VS_control"),
color_label = "both_up",label_size = 4)
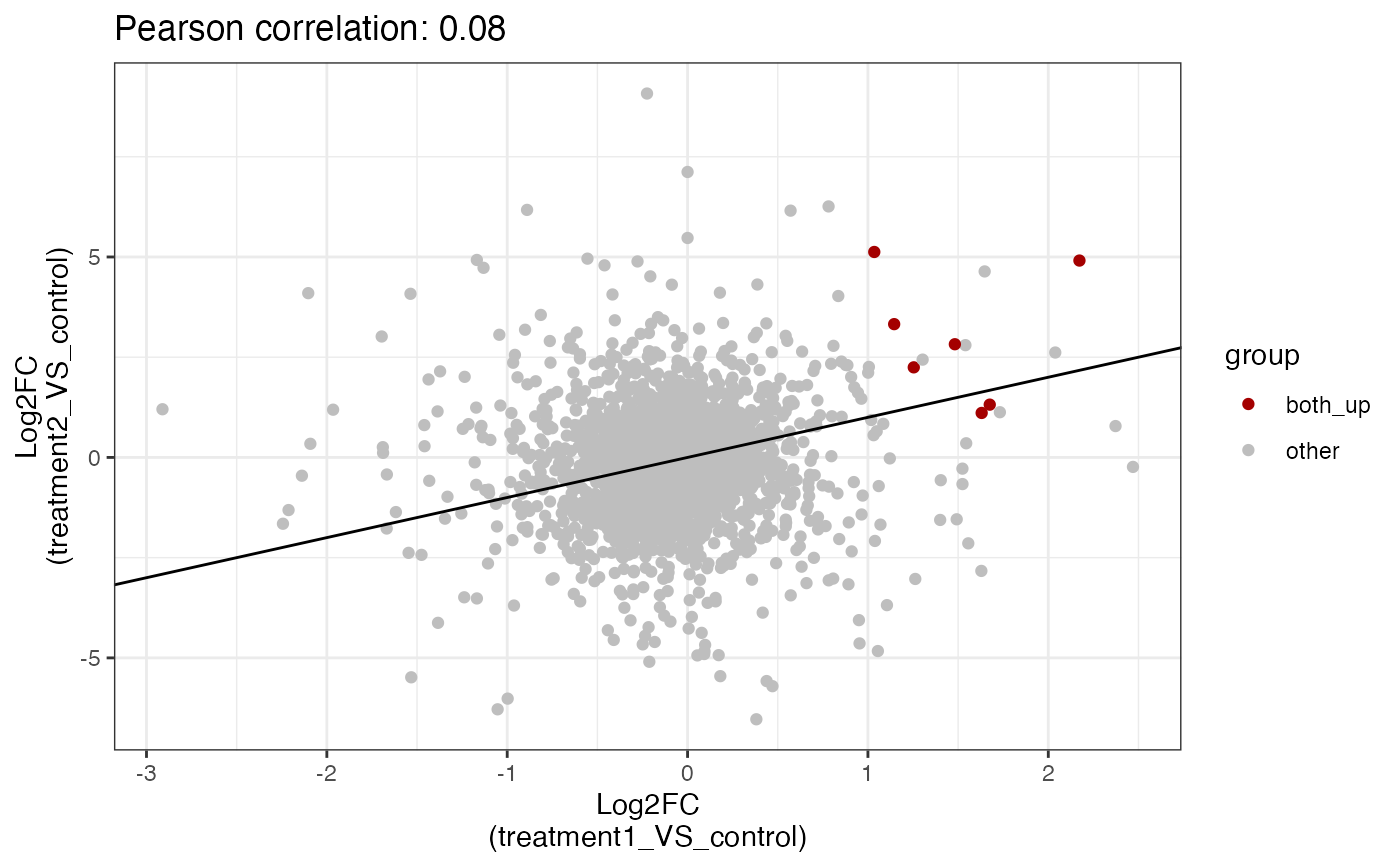 # show common down
get_fold_change_scatter_plot(x = res,
sample_comparisons = c("treatment1_VS_control",
"treatment2_VS_control"),
color_label = "both_down",label_size = 4, point_size = 4)
# show common down
get_fold_change_scatter_plot(x = res,
sample_comparisons = c("treatment1_VS_control",
"treatment2_VS_control"),
color_label = "both_down",label_size = 4, point_size = 4)