Plot a venn diagram for diff ASE (using event names or gene names) between multiple comparison.
Source:R/splicewiz_wrappers.R
plot_deASE_venn.RdPlot a venn diagram for diff ASE (using event names or gene names) between multiple comparison.
Usage
plot_deASE_venn(
x,
sample_comparisons,
regulation = "both",
fill_color = NULL,
event_type = NULL,
ovelap_genes = TRUE,
...
)Arguments
- x
an abject of class "parcutils_ase". This is an output of the function
run_ase_diff_analysis().- sample_comparisons
a character vector denoting sample comparisons for which ASE to be obtained.
- regulation
a character string, default
both. Values can be one of theup,down,both,other,all.- fill_color
an argument pass to the function ggvenn::ggvenn.
- event_type
a character string denoting event type for which one of the
up,down,both,otherorallevents to be returned based. Choices are:IR,SE,AFE,ALE,A3SS,A5SSandMXE.Default NULL, uses all event types.- ovelap_genes
logical, default
TRUE, instead of event_names, overlap gene names.- ...
other arguments pass to the function ggvenn::ggvenn
Examples
se <- SpliceWiz::SpliceWiz_example_NxtSE(novelSplicing = TRUE)
SpliceWiz::colData(se)$treatment <- rep(c("A", "B"), each = 3)
SpliceWiz::colData(se)$replicate <- rep(c("P","Q","R"), 2)
res <- run_ase_diff_analysis(x = se, test_factor = "replicate", test_nom = c("P","Q","R") ,test_denom = c("Q","R","P"), IRmode ="annotated", cutoff_lfc = 0.6, cutoff_pval = 1, regul_based_upon = 1)
#> Aug 16 11:35:36 Performing edgeR contrast for included / excluded counts separately
#> Aug 16 11:35:37 Performing edgeR contrast for included / excluded counts together
#> Aug 16 11:35:42 Performing edgeR contrast for included / excluded counts separately
#> Aug 16 11:35:43 Performing edgeR contrast for included / excluded counts together
#> Aug 16 11:35:47 Performing edgeR contrast for included / excluded counts separately
#> Aug 16 11:35:48 Performing edgeR contrast for included / excluded counts together
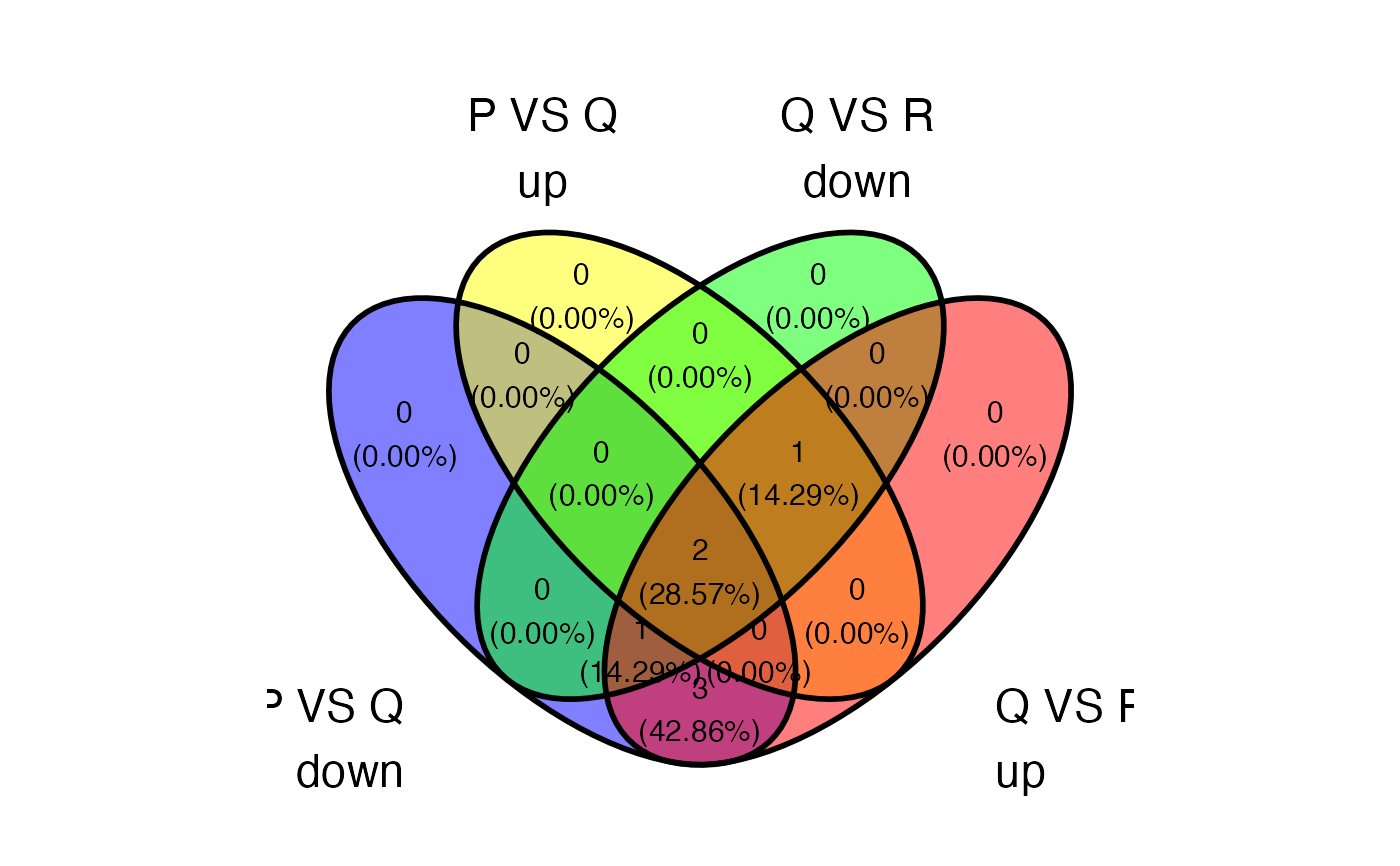
plot_deASE_venn(x = res, sample_comparisons = c("P_VS_Q","Q_VS_R","R_VS_P"), event_type = "SE", regulation = "both", ovelap_genes = TRUE)
#> ! Maximum 4 sets can be included in the venn diagram. These sets won't be included: R_VS_P_down and R_VS_P_up
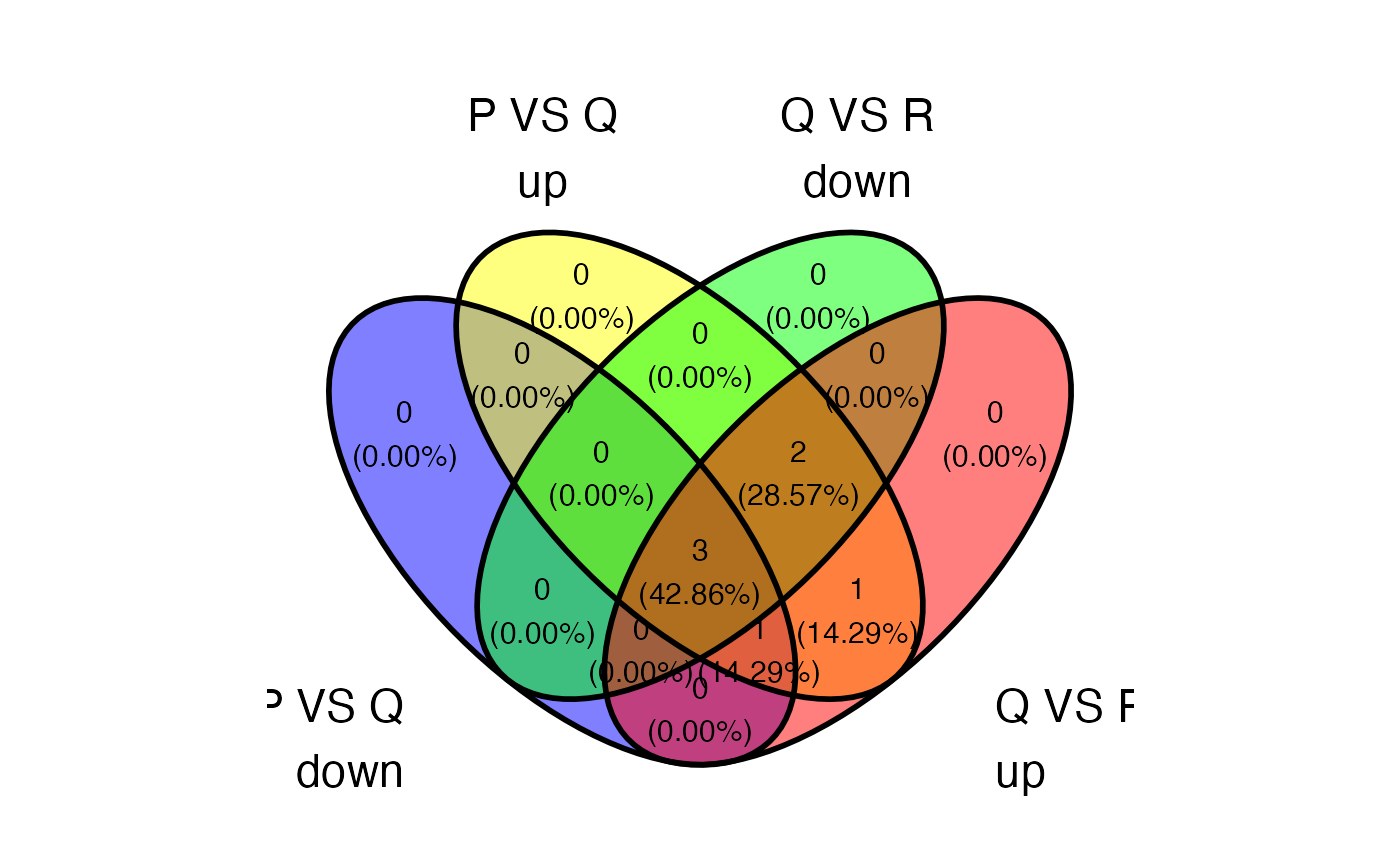 plot_deASE_venn(x = res, sample_comparisons = c("P_VS_Q","Q_VS_R","R_VS_P"), event_type = "SE", regulation = "up", ovelap_genes = TRUE)
plot_deASE_venn(x = res, sample_comparisons = c("P_VS_Q","Q_VS_R","R_VS_P"), event_type = "SE", regulation = "up", ovelap_genes = TRUE)
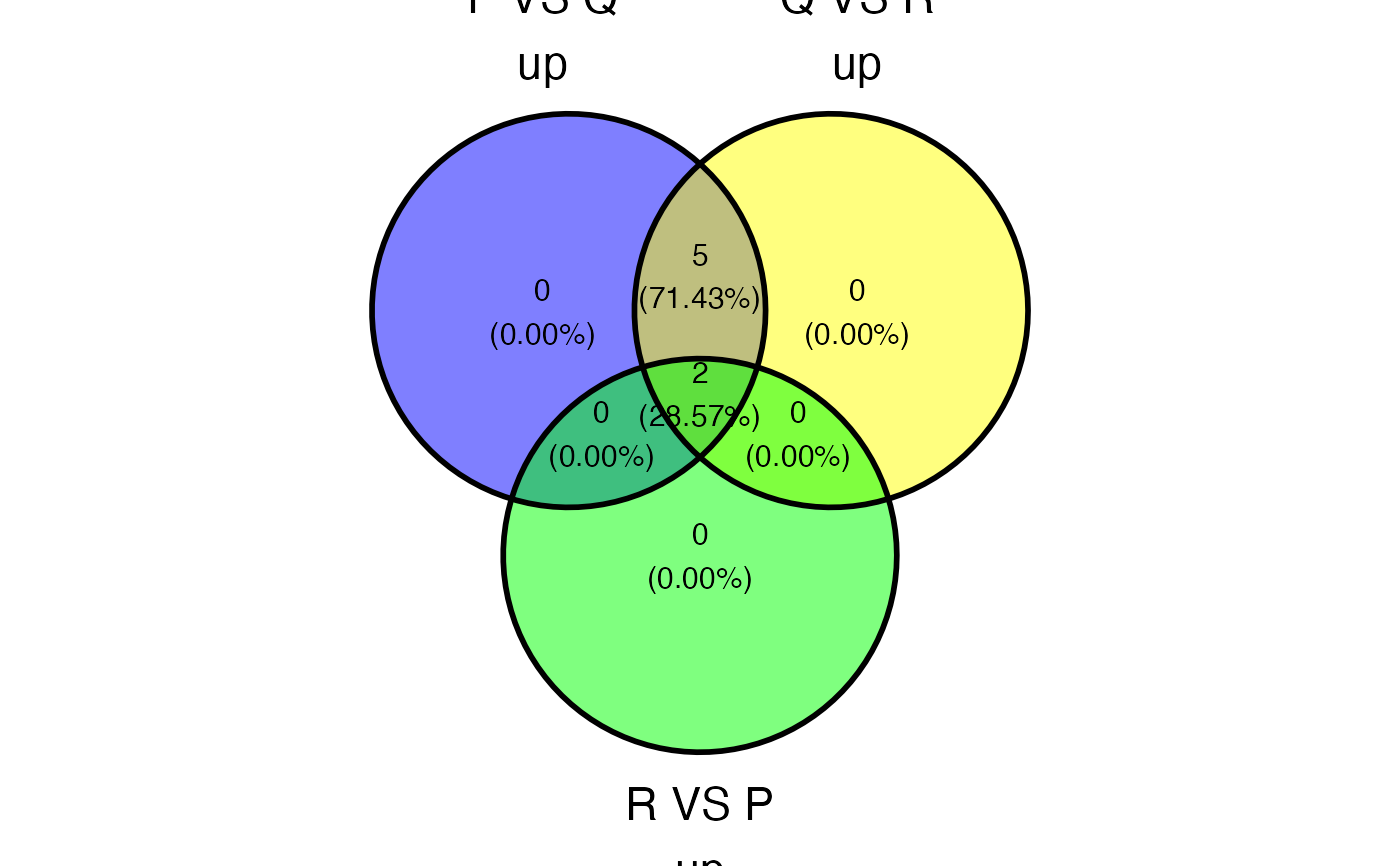 plot_deASE_venn(x = res, sample_comparisons = c("P_VS_Q","Q_VS_R","R_VS_P"), event_type = "A3SS", regulation = "both", ovelap_genes = TRUE)
#> ! Maximum 4 sets can be included in the venn diagram. These sets won't be included: R_VS_P_down and R_VS_P_up
plot_deASE_venn(x = res, sample_comparisons = c("P_VS_Q","Q_VS_R","R_VS_P"), event_type = "A3SS", regulation = "both", ovelap_genes = TRUE)
#> ! Maximum 4 sets can be included in the venn diagram. These sets won't be included: R_VS_P_down and R_VS_P_up